gpDB: a database for
G-proteins GPCRs and their interaction

User’s Manual
Theodoropoulou, M.C., Elefsinioti, A.L., Bagos, P.G., Spyropoulos, I.C.
and
Hamodrakas, S.J.
Downloaded from http://bioinformatics.biol.uoa.gr/gpDB
Basic theory
G-proteins
act as switches for signal transduction from extracellular space into the
cell. This is accomplished through
their interaction with G-Protein Coupled Receptors (GPCRs). G-proteins form hetero-trimers composed of Gá,
Gâ and Gă
subunits, and they also possess a binding site for a nucleotide (GTP or
GDP). G-proteins are named after their á-subunits,
which on the basis of their amino acid similarity and function are grouped into
four families (Gás, Gái/o, Gáq, Gá12).
GPCRs form the major group
of receptors in eukaryotes and they possess seven transmembrane á-helical domains. GPCRs are usually classified
into several classes, according to the sequence similarity shared by the
members of each class.Class A of GPCRs (rhodopsin-like GPCRs) contains the
majority of GPCRs, including receptors for structurally diverse ligands
(biogenic amines, nucleotides, peptides, glycoprotein hormones etc). Class B (secretin-like GPCRs) contains
purely peptide receptors, whereas class C (metabotropic glutamate family
receptors) contains metabotropic glutamate and GABA-B receptors and some taste
receptors. Class D contains the fungal pheromone receptors, class E contains
the cAMP receptors of Dictyostelium and last is the Frizzled/Smoothened
class. There is also a number of
putative classes of newly discovered GPCRs, whose nomenclature has not been
accepted yet from the scientific community.
The
stimulation of GPCRs leads to the activation of G-proteins, which dissociate
into Galpha and Gbeta-gamma subunits. The subunits then activate several
effector molecules that lead to many kinds of cellular and physiological
responses.
Effectors form a diverse
group of proteins, that, throught their interaction with G-proteins, either act
as second messengers, or lead directly to a cellular and physiological
response. Effectors have never being classified before. We classified them into
families, subfamilies and types, based on their function.
Data
annotation
The annotation regarding the interaction between
GPCRs, G-proteins and effectors and the effect of the particular interaction was
a result of an exhaustive and detailed literature search. We collected the
available information from review articles and original research papers, which
we provide as links in each entry page. A point that it was impossible to be
explained in the manuscript (and thus is discussed in the online manual pages)
is the fact that no entries are included in the database solely using a
prediction system. On the contrary, interactions are inferred from
orthologues. In particular, when we
have a particular reference stating that protein X interacts with protein Y in
organism Z, we search all the other closely related organisms for such pairs
(X-Y). This search is not being performed on an automated fashion (i.e. a
simple BLAST search) but instead we rely on family classification (from PFAM),
the gene name, the function of the proteins etc.
For instance from the reference that used fused chimeric mutants of bovine ACI:
“Wittpoth C, Scholich K, Yigzaw Y, Stringfield TM, Patel TB. Regions on adenylyl cyclase that are necessary for inhibition of activity by beta gamma and G(ialpha) subunits of heterotrimeric G proteins. Proc Natl Acad Sci U S A. 1999;96(17):9551-6.”
we conclude that Gbeta-gamma dimer inhibits Adenylyl
Cyclase I, and thus this information could be transferred to all the available
(mostly mammalian) organisms possessing Gbeta-gamma and ACI.
From the paper describing another heterologous
expression system:
“Marty C,
Browning DD, Ye RD. Identification of tetratricopeptide repeat 1 as an adaptor
protein that interacts with heterotrimeric G proteins and the small GTPase Ras.
Mol Cell Biol. 2003;23(11):3847-58”
we conclude that Galpha-16 interacts with TRP1 and
this information could be expanded to all organisms possessing Galpha-16 and
TRP1. And similarly we proceed with the other interactions.
From the paper describing the selectivity of AT2
receptor in the Rat fetus:
“Zhang
J, Pratt RE. The AT2 receptor selectively associates with Gialpha2 and Gialpha3
in the rat fetus. J Biol Chem. 1996 Jun 21;271(25):15026-33“
we
conclude that AT2 receptor interacts with Galpha-I and this information could
be transferred to all organisms possessing AT2 receptor and Galpha-i.
From the paper describing another heterologous
expression system:
“Borowsky
B, Adham N, Jones KA, Raddatz R, Artymyshyn R, Ogozalek
KL, Durkin MM, Lakhlani PP, Bonini JA, Pathirana S,
Boyle N, Pu X, Kouranova E, Lichtblau H, Ochoa
FY, Branchek TA, Gerald C. Trace amines:
identification of a family of mammalian G protein-coupled receptors. Proc Natl Acad Sci U S A.
2001 Jul 31;98(16):8966-71”
we
conclude that TA1 receptor couples with Galpha-s and this information could be
expanded to all organisms possessing TA1 and Galpha-s.
Of
course, there are other more “simple” and straightforward situations such as
the interactions of Galpha-s subunits that are known for years to stimulate
adenylate cyclases, and so on.
This
way, although someone could argue that some of these entries should be marked “by
similarity” we feel that we should not use such discrimination in the
annotation of the database entries.
Scope of
the gpDB
GpDB is a publicly
accessible, relational
database,
containing information about G-proteins, GPCRs and Effectors. It contains
detailed information for 391 G-proteins (250 G-alpha, 84 G-beta and 57
G-gamma), 2738 GPCRs belonging to families with known coupling to G-proteins,
and 1390 Effectors, that interact with specific G-proteins. The sequences are classified according to a hierarchy of
different classes, families and sub-families, based on literature search. Particularly, effectors are classified
into families, subfamilies and types. The main innovation besides the
classification of G-proteins, GPCRs and effectors is the relational model of
the database, describing the known coupling specificity of the GPCRs to their
respective alpha subunit of G-proteins and also the specific interaction
between the different subfamilies of G-proteins and particular effector types,
a unique feature not available in any other database. There is full sequence
information with cross-references to publicly available databases, and the user
may submit advanced queries for text search.
Furthermore there is interconnectivity with PRED-GPCR, PRED-TMR,
TMRPres2D, a pattern search tool, and an interface for running BLAST against
the database. The database will be very useful for the study of G-protein/GPCR
and G-protein/Effectors interactions, and for future development of algorithms
predicting this interaction. It can be accessed via a web-based browser at the
URL: http://bioinformatics.biol.uoa.gr/gpDB
Navigation
Through the
navigation tool, the user has the ability to browse the database following the
hierarchy. The navigation could be performed on the GPCRs, the G-PROTEINS or
the EFFECTORS hierarchy. Following the link of GPCRs, the user may be navigated
through:
GPCR CLASSES
Top of the GPCR classes page
GPCR
FAMILIES
We
have classified GPCRs into 64 different families

Top of the
GPCR families page
GPCR SUB-FAMILIES
Each family is further
subdivided into different subfamilies, based mainly on TIPS classification
scheme that takes into account the native ligand(s) that binds to a particular
GPCR .
The GPCR SUBFAMILIES MENU
enables the user to either view the individual receptors of the specific
subfamily or to view the coupling specificity of the GPCR subfamily with
G-protein subfamilies.
|
Viewing the
receptors of the specific subfamily By clicking on a specific
subfamily the user is presented with a list of all individual receptors
belonging to this subfamily EXAMPLE The user is able to click
on a specific subfamily like 5-HT Subfamily of the
5-HYDROXYTRYPTAMINE RECEPTOR
Subfamilies of 5-Hydroxytryptamine
receptor family The result page presents
all the individual receptors of 5-HT subfamily
Receptors of 5-HT
subfamily |
Coupling
between GPCRs - G-protein subfamilies The user has the
potential to see the coupling specificity of a GPCR subfamily with G-proteins
subfamilies EXAMPLE As it is shown in the
picture of the GPCR subfamilies the user has the alternative to click on the arrow button instead of selecting to
click on a specific subfamily. The user then is
presented with a list of G-protein subfamilies that couple to the specific
GPCR subfamily. The G-protein types of these subfamilies have known coupling
specificity to the receptors of this specific GPCR subfamily.
The result page |
Following the link of
G-PROTEINS, the user may browse through:
G-PROTEIN CLASSES

G-PROTEIN FAMILIES
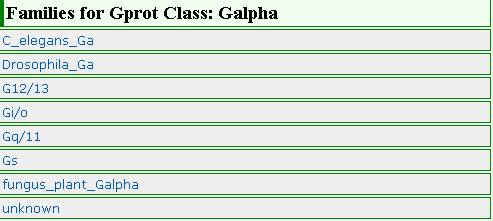
Families of Galpha class
G-PROTEIN
SUB-FAMILIES
The G-PROTEIN SUBFAMILIES MENU enables the user to either view the
protein types of the specific subfamily or to view the coupling specificity of
the G-protein subfamily with GPCR subfamilies.
|
Viewing the
types of the specific subfamily By clicking on a specific
subfamily the user is presented with a list of all G-protein types belonging
to this subfamily EXAMPLE The user is able to click
on a specific subfamily like Galpha-12/13 subfamily of Gi/o family
Subfamilies of G12/13
family The result page presents
all the G-protein types of Galpha-12 subfamily G-PROTEIN TYPES
Types of Galpha-12
subfamily Then by
clicking on any type the user is presented with all individual G-proteins Ending up to
individual G-proteins.
Proteins
of Galpha-12 type |
Coupling
between GPCRs -G-proteins subfamilies The user has the potential
to see the coupling specificity of a G-protein subfamily with GPCR
subfamilies EXAMPLE As it is shown in the
picture of the G-protein subfamilies the user has the alternative to click on
one of the two arrow buttons
instead of selecting to click on a specific subfamily.
Subfamilies of Gi/o
family The red button
presents the user a list of GPCR subfamilies that couple to the
specific G-protein subfamily. The GPCRs of these subfamilies have known coupling
specificity to the receptors of this specific G-protein subfamily
The result page The yellow button presents the user a list of effector types with whom this
G-protein subfamily interacts.
The result page |
Following the link of
Effectors, the user may browse through:
EFFECTOR FAMILIES
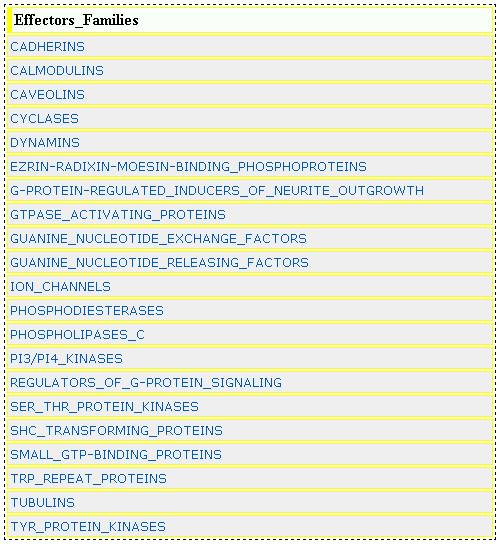
Effector Families
EFFECTOR SUB-FAMILIES
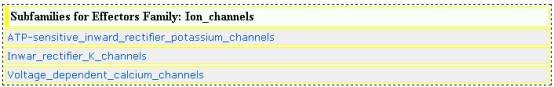
Subfamilies of ion channels family
EFFECTOR
TYPES
The Effector TYPES MENU enables the user to either view the entries of
this protein type or to view the G-protein subfamilies, that interact with this type.
|
Viewing the
entries of the specific type By clicking on a specific
type the user is presented with a list of all Effector types belonging to
this subfamily EXAMPLE The user is able to click
on a specific type like ATP-sensitive
inward rectifier potassium channel-1
type of the ATP-sensitive inward rectifier
potassium channel subfamily
Types of ATP-sensitive
inward rectifier potassium channels subfamily By clicking on a
type the user is presented with all individual Effectors Ending up to
individual Effectors.
Proteins
of ATP-sensitive inward
rectifier potassium channel-1 type |
Interaction
between G-proteins subfamilies and Effector types The user has the
potential to see the interaction between G-proteins subfamilies and Effector
types EXAMPLE As it is shown in the picture
of the Effector types the user has the alternative to click on the arrow button instead of selecting to
click on a specific type.
Types of ATP-sensitive
inward rectifier potassium channels subfamily The user then is
presented with a list of G-protein subfamilies that interact with this
particular Effector type.
The result page |
At each point the user may
navigate up or down to the hierarchy tree.
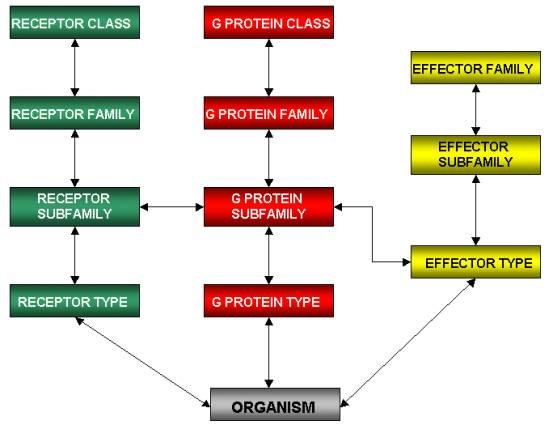
Figure
1. The relational model of the database
Text
Search
In the Text Search area,
the user can search for any text in the fields of his/her preference. The user
can enter any word in one or more of the available boxes under the name: 'Protein
Name', 'Species', ’Common Name’, 'Description', 'Gene
Name' and 'Cross-References'. The user has also the ability to
select if he wants to exclude fragments from the results.
Each expression may
contain:
i) Text terms to be searched for,
ii) Parenthesis '(' ')' which groups one or more sub-expressions,
iii) Operator '&' for AND, which combines two (or more)
sub-expressions in a single field and gives the user the opportunity to search
for entries that satisfy all sub-expressions.
iv) Operator '|' for OR, which combines two (or more)
sub-expressions in a single field and gives to the user the opportunity to
search for entries that satisfy at least one of the sub-expressions.
v) Operator '!' for NOT, which can only be used at the beginning of
an expression and does not connect two sub-expressions. It provides to the user
the opportunity to search for entries that necessarily do not satisfy the
expression after the '!'.
vi) Operator '&!' for AND NOT, which combines two (or more)
sub-expressions in a single search field and gives to the user the opportunity
to search for entries that satisfy only the sub-expression in the left of the
operator.
Expressions in
separate search fields are combined with the AND operator, so every entry of
the result set will satisfy the expressions of all the search fields the user
has chosen. The user has the option to choose whether the query will be
performed against the GPCRs or the G-Proteins included in the database.
PROTEIN NAME
Corresponds
to the field PROTEIN NAME of an entry
EXAMPLE
If the user wants to retrieve all Galpha proteins
he/she has to use the name “Galpha” as a query in the PROTEIN NAME box
and select G-protein from Search Target field.
The
top of the result page is:
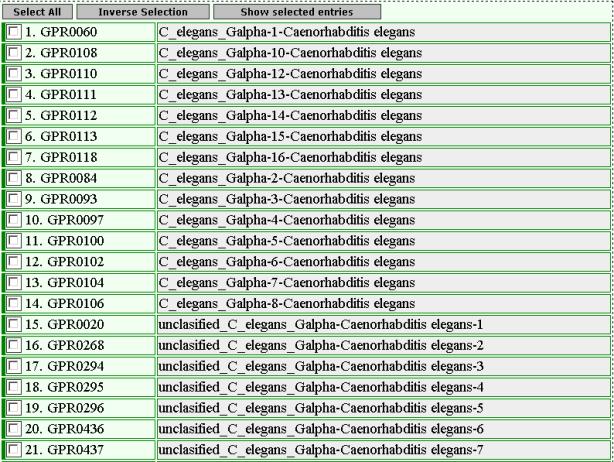
The user has to use the name “Gbeta” and “Ggamma”
in order to search for Gbeta and Ggamma proteins additionally
SPECIES
Corresponds
to the field SPECIES of an entry (the scientific name of a species)
EXAMPLE
1)
If the user wants to retrieve all G-proteins of Drosophila he/she has to
use the name “Drosophila” as a query in the SPECIES box and
select G-protein from Search Target field.
The result
page is:
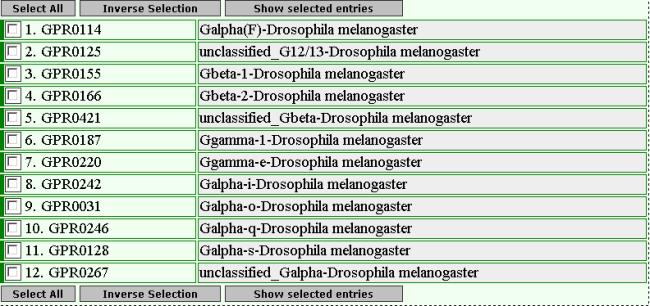
2)
If the user wants to retrieve all GPCRs of Drosophila he/she has to use
the name “Drosophila” as a query in the SPECIES box and select GPCR
from Search Target field.
The top of the result page is:

COMMON NAME
Corresponds
to the field COMMON NAME of an entry (the common name of a species)
EXAMPLE
3)
If the user wants to retrieve all G-proteins of Drosophila melanogaster
he/she has the alternative to use the name “fruit fly” (The common name
of Drosophila melanogaster) as a query in the COMMON NAME box and select
G-protein from Search Target field.
The result page is:

REFERENCES
Corresponds
to the field DESCRIPTION of an entry
The
user is able to use any accession numbers and/or IDs from other databases such
as SWISS_PROT, PIR, MIM, PRODOM, GENEW, PRINTS, INTERPRO etc.
EXAMPLE
If the user wants to retrieve a G-protein that has “P29348”
as an accession number in SWISS_PROT he has to use “P29348” as a query
and select G-protein from Search Target field.
The
top of the result page is:

DESCRIPTION
Corresponds
to the field DESCRIPTION of an entry
EXAMPLE
The
top of the result page is:
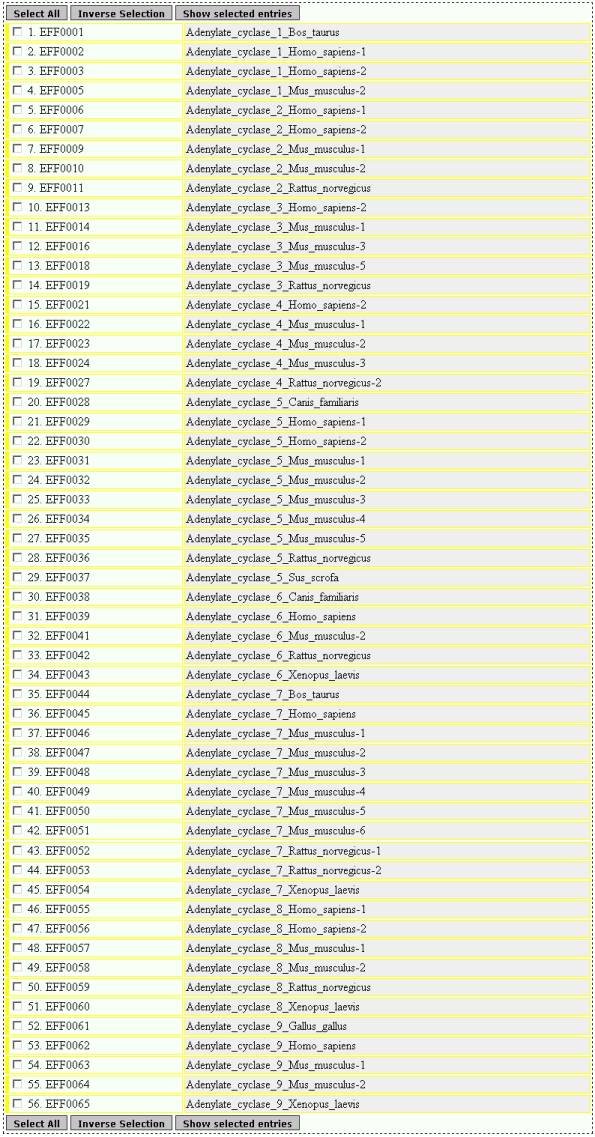
GENE NAME
Corresponds to the field GENE of an entry
EXAMPLE
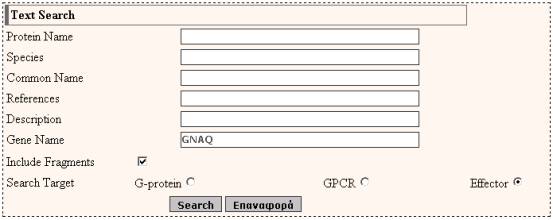
The
result page is:
BLAST
Search
With the BLAST search tool,
the user may submit a sequence and search the database for finding homologues.
The user has the option to choose whether to perform the BLAST search against
GPCRs sequences, G-proteins sequences and/or Effectors sequences. The input for
the BLAST application is the sequence in standard FASTA format.
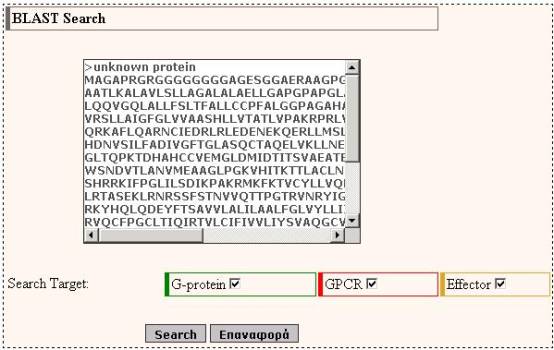
Submitting a sequence
The output of the BLAST
query consists of a list of sequences in the database having significant
E-values in a local pairwise alignment, ranked by statistical significance. In
the output, are also listed the range of residues in which the alignment occurs,
in both the target and the query sequence, the number of identical and similar
residues in the alignment and the E-value of the alignment.
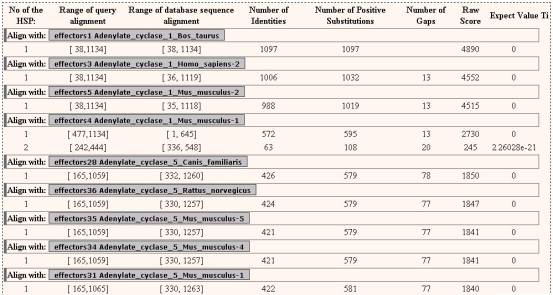
By clicking the NAME
button from each hit, the user may visualize the local alignment
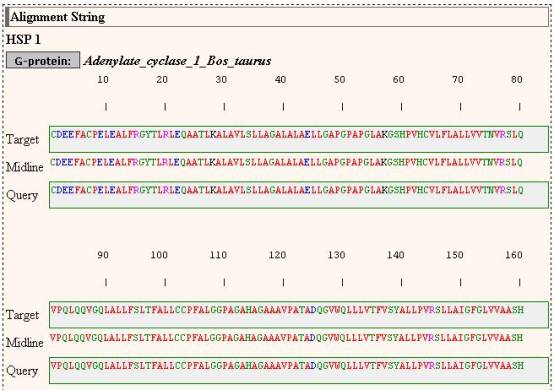
The top of the result page
And from there, the user
may retrieve the detailed view of the entry corresponding to the
particular target sequence.
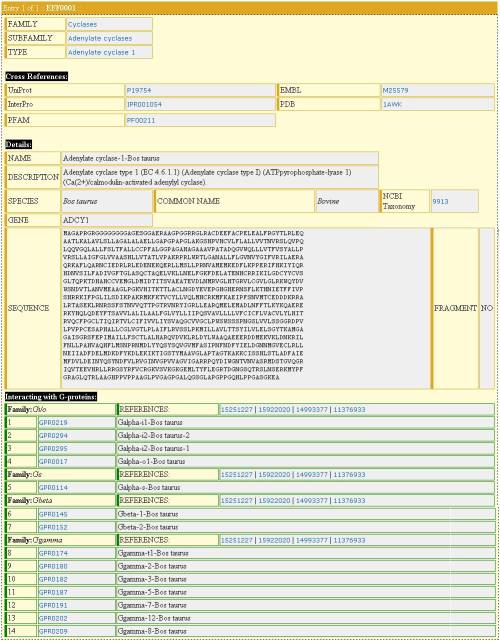
![]() The entry of the particular target sequence
The entry of the particular target sequence
![]()
Pattern
Search
Using the
Pattern Search tool, the user may perform searches for finding specific
patterns in the proteins of the database. The user has the option to choose
whether to perform the Pattern search against the GPCRs sequences or the
G-Proteins. The input of the Pattern
Search tool is a regular expression pattern following the PROSITE syntax.
The output of
the Pattern search application consists of a list of the sequences matching the
particular pattern. gpDB ID(s) and the NAME of the target sequence(s) are
listed in the output. The user has the option to check the entry or the entries
that he/she wants to retrieve, and after pressing the appropriate button, to
have them in the detailed view.
Detailed
View of a Protein
The detailed view of an
entry corresponds to the last level of the hierarchy. In the detailed view, the
available information regarding a GPCR, a G-protein or an Effector sequence is
presented.
The fields of the detailed
view are the following :
- Classification of the particular protein according
to the hierarchy
- Gpdb Name
- Gpdb ID
- The gene name
- The scientific name of the species that the
protein was found
- The common name of the species that the protein
was found
- The sequence
- A short description
- Available links with cross-references to the
publicly available databases (UNIPROT, PIR, PDB, InterPro, PRINTS, PFAM,
MIM, NCBI Gene etc)
- INFORMATION ABOUT DIMERIZATION (only GPCRs)
- Scaffold proteins that couple to the particular
GPCR subfamily
- Finally and more important, there is a
list with the available information regarding the coupling
specificity between GPCRs and G-proteins and/or the interaction
between G-proteins and Effectors.
Additionally
GPCR entries have links to PRED-GPCR,
PRED-COUPLE2
and HMM-TM
PRED-GPCR,
PRED-COUPLE2 and HMM-TM are tools that were developed in our laboratory.
PRED-GPCR is a system based on a probabilistic method
that uses family specific profile HMMs in order to determine to which GPCR
family a query sequence belongs or resembles.
PRED-COUPLE2 is a system based on a refined library of highly-discriminative Hidden
Markov Models in order to predict the coupling specificty of GPCRs to all
families of G-proteins (including G12/13). Hits from individual profiles are
combined by a feed-forward Artificial Neural Network to produce the final
output.
HMM-TM is an algorithm for the
prediction of the topology of transmembrane proteins using HMMs.
By clicking on the
representation button the user gains access to another tool of our laboratory TMRPres2D
The 'TransMembrane protein Re-Presentation
in 2 Dimensions' tool, automates the creation of uniform, two-dimensional,
high analysis graphical images/models of alpha-helical or beta-barrel
transmembrane proteins
The gpDB accession numbers
and the names of the proteins with which that protein couples, are listed with
the appropriate links. By clicking on any of these links, the user will
be presented with the detailed view of the corresponding protein.
The detailed view, of
GPCRs, G-Proteins and Effectors are completely analogous, with the only
difference being the fact that the coupling relationship is of the type
“many-to-many”. This means, that a particular G-protein, may couple to
more than one receptor of the same organism (which is usually the case) but
that particular GPCR may also couple to other G-proteins of the same
organism (promiscuous coupling). This also happens between G-Proteins
and Effectors. Especially for GPCRs, the user has also the option to submit
their sequences to the PRED-GPCR server and retrieve prediction regarding the
classification of the receptor
EXAMPLE
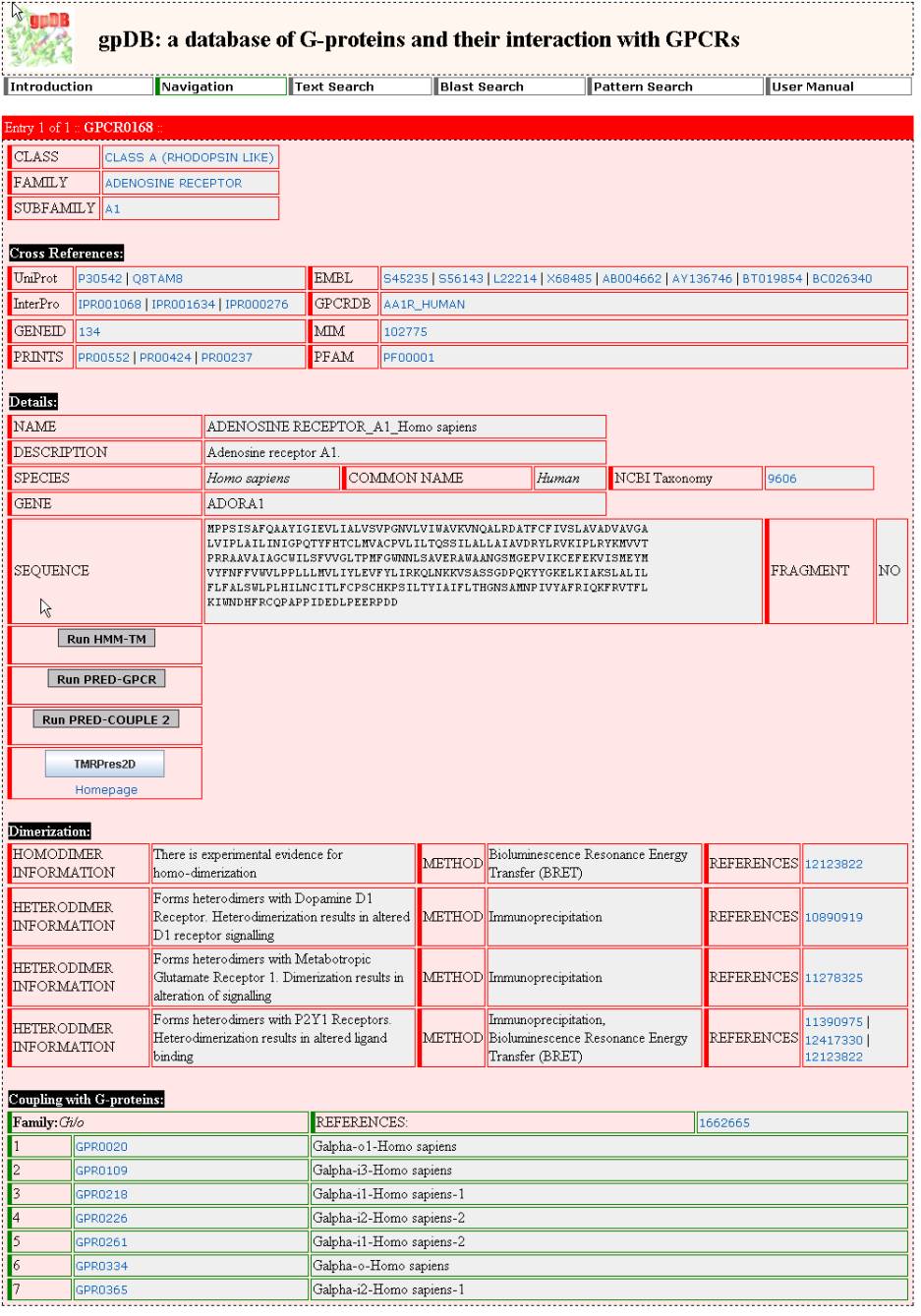
 Complete entry of A1 Adenosine receptor of Homo
sapiens.
Complete entry of A1 Adenosine receptor of Homo
sapiens.